https://doi.org/10.1140/epjd/s10053-025-01024-8
Regular Article – Quantum Information
Quantum drug discovery: a hybrid quantum graph neural network–variational quantum eigensolver framework for serine neutralization
1
Fakultät Wirtschaft, Hochschule Heilbronn, Max-Planck-Str.39, 74081, Heilbronn, Baden-Württemberg, Germany
2
Department of Mechanical Engineering, Universidad de La Rioja, Edificio Departamental, c/ San José de Calasanz, 31, 26004, Logroño, La Rioja, Spain
a
javier.villalba-diez@hs-heilbronn.de
Received:
5
April
2025
Accepted:
8
June
2025
Published online:
11
July
2025
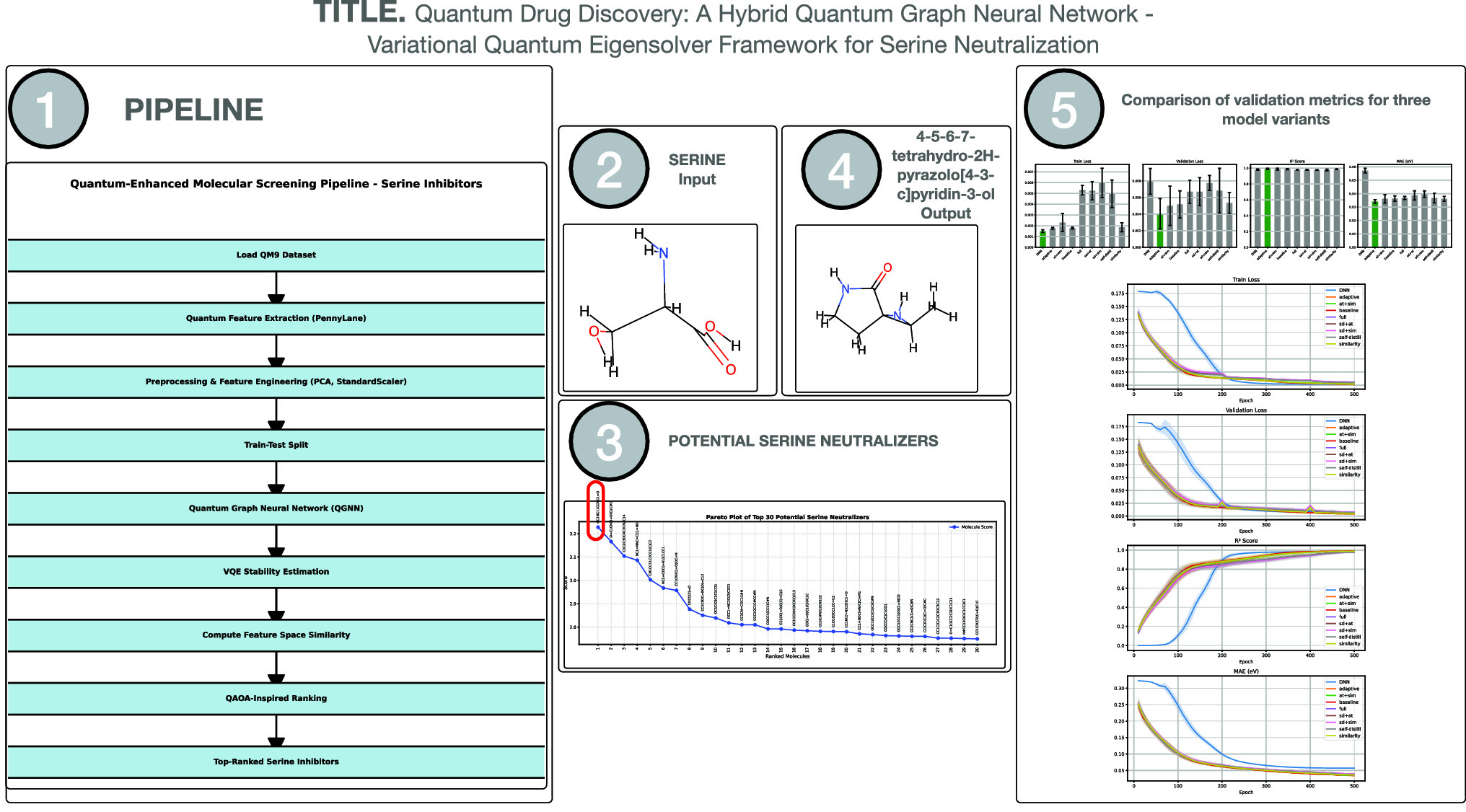
Quantum computing has emerged as a powerful tool for modeling molecular systems with high accuracy, accelerating the search for novel therapeutic targets. In this study, we present a two-stage hybrid workflow that combines a Quantum Graph Neural Network with a Variational Quantum Eigensolver to identify potent serine neutralizers in the QM9 dataset. In the first stage, an advanced quantum graph neural network architecture incorporating attention layers, self-distillation, and an adaptive learning-rate schedule is trained to predict ionization potentials and binding free energies. Across five independent random-seed trials, our adaptive-thresholded QGNN–VQE pipeline achieves an average 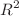 of
of 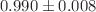 and a mean absolute error of
and a mean absolute error of  eV (
eV (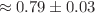 kcal/mol) on the QM9 validation set, demonstrating robust, chemical-accuracy-level predictions. Building on these high-precision predictions, the second stage employs a QAOA-inspired hybrid ranking scheme that merges quantum graph neural network outputs, feature-space similarity (via PCA and cosine similarity), and variational quantum eigensolver-derived energy stability. This
kcal/mol) on the QM9 validation set, demonstrating robust, chemical-accuracy-level predictions. Building on these high-precision predictions, the second stage employs a QAOA-inspired hybrid ranking scheme that merges quantum graph neural network outputs, feature-space similarity (via PCA and cosine similarity), and variational quantum eigensolver-derived energy stability. This 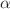 -weighted (
-weighted (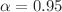 ) scoring framework is applied to over 133,000 molecules, efficiently parsed in parallel. The final ranked list is validated through an improved bijection test, featuring an adaptive similarity threshold to ensure both structural and functional robustness. The top-scoring candidate identified by our pipeline is 5,6,7-tetrahydro-4 H-pyrazolo[4,3-c]pyridin-4-one, highlighting the efficacy of quantum-enhanced modeling in pinpointing complex pharmacological targets. These findings underscore the transformative potential of hybrid quantum-classical workflows in drug discovery, offering a flexible blueprint for advanced screening of other critical biomolecular interactions in the rapidly expanding field of quantum biology.
) scoring framework is applied to over 133,000 molecules, efficiently parsed in parallel. The final ranked list is validated through an improved bijection test, featuring an adaptive similarity threshold to ensure both structural and functional robustness. The top-scoring candidate identified by our pipeline is 5,6,7-tetrahydro-4 H-pyrazolo[4,3-c]pyridin-4-one, highlighting the efficacy of quantum-enhanced modeling in pinpointing complex pharmacological targets. These findings underscore the transformative potential of hybrid quantum-classical workflows in drug discovery, offering a flexible blueprint for advanced screening of other critical biomolecular interactions in the rapidly expanding field of quantum biology.
© The Author(s) 2025
 Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.