https://doi.org/10.1140/epjd/e2002-00157-4
Toward direct determination of conformations of protein building units from multidimensional NMR experiments III
A theoretical case study of For–L-Phe–NH
1
Department of Organic Chemistry, Eötvös University, P.O. Box 32,
1518 Budapest 112, Hungary
2
Department of Theoretical Chemistry, Eötvös University, P.O. Box 32,
1518 Budapest 112, Hungary
Corresponding authors: a perczel@para.chem.elte.hu - b csaszar@chem.elte.hu
Received:
10
January
2002
Published online:
13
September
2002
Chemical shielding anisotropy tensors have been determined, within the GIAO-RHF formalism
using a smaller [6-31+G(d)] and two medium-size basis sets [6-311++G(d,p) and TZ2P], for all
elements of the conformational library (altogether 27 structures) of the hydrophobic model peptide For–L-Phe–NH2. The individual chemical shifts and their conformational averages have been compared to
their experimental counterparts taken from the BioMagnetic Resonance Bank (BMRB). At the highest
level of theory applied, for all nuclei but the amide proton, deviations between statistically averaged
theoretical and experimental chemical shifts are as low as a few percent. One-dimensional (1D)
chemical shift – structure plots do not allow unambiguous identification of backbone conformations. On
the other hand, on chemical shift – chemical shift plots of selected nuclei, e.g., 1HN with 15N or 15N with
13C , regions corresponding to major conformational motifs have been found, providing basis for the
identification of peptide conformers solely from NMR shift data. The 2D 1H
, regions corresponding to major conformational motifs have been found, providing basis for the
identification of peptide conformers solely from NMR shift data. The 2D 1H –13C
–13C as well as the 3D
1H
as well as the 3D
1H –13C
–13C –13C
–13C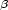 chemical shift – chemical shift plots appear to be of special importance for direct
determination of conformations of protein building units from multidimensional NMR experiments. 48
pairs of 1H
chemical shift – chemical shift plots appear to be of special importance for direct
determination of conformations of protein building units from multidimensional NMR experiments. 48
pairs of 1H –13C
–13C data for phenylalanine residues have been extracted from 18 selected proteins and
compared to relevant ab initio results, supporting the calculated results. Thus, the appealing idea of
establishing backbone folding information of peptides and proteins from chemical shift information
alone, obtained from selected multiple-pulse NMR experiments (e.g., 2D-HSQC, 2D-HMQC, and
3D-HNCA), has received further support.
data for phenylalanine residues have been extracted from 18 selected proteins and
compared to relevant ab initio results, supporting the calculated results. Thus, the appealing idea of
establishing backbone folding information of peptides and proteins from chemical shift information
alone, obtained from selected multiple-pulse NMR experiments (e.g., 2D-HSQC, 2D-HMQC, and
3D-HNCA), has received further support.
PACS: 31.15.Ar – Ab initio calculations / 33.25.+k – Nuclear resonance and relaxation
© EDP Sciences, Società Italiana di Fisica, Springer-Verlag, 2002





